Thematics Genomes and Evolution
Impact of selfish DNA and virus on genome evolution
Selfish DNA refers to genomic entities that are able to transmit more efficiently than the rest of the genome to the next generation, regardless of their potentialy deleterious effects to the host. By this, they are source of intragenomic conflicts. They include transposable elements (TEs), meiotic distorters and B chromosomes.
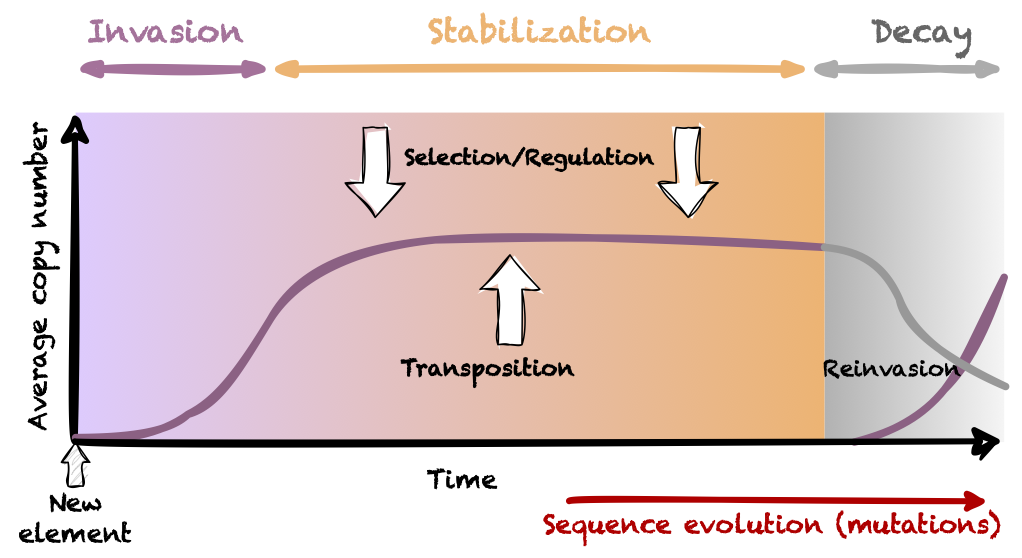
Transposable elements are DNA mobile, repeated and dispersed sequences and able to invade genomes as well as populations. They are present in all genomes, of which they make a significant part. Because TE copies have no function in the cell, a TE family will evolve free of selection within a species, and will accumulate mutations leading to the inactivation of the family. Furthermore, genome defense involving small RNA can silence TEs and prevent transposition. The persistence of active families is explained by recurrent horizontal transfers of some copies between species.
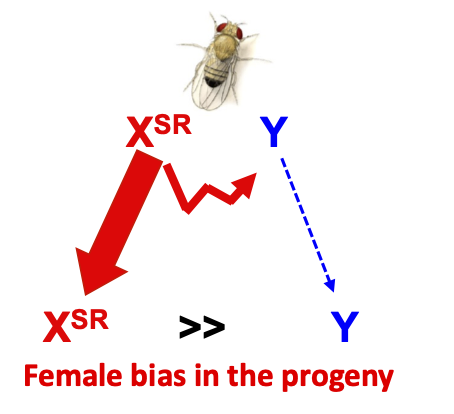
Meiotic distorters are loci that do not follow mendelian segregation, and are preferentially transmitted to the next generation (relative to the homologous non distorter allele). In XY system, distorters on the X such as the Paris element, highly bias the progeny of the carrier male toward females. Restoration of equilibrated sex-ratio involve the emergence of autosomal suppressors or resistant Y chromosomes. Other entities behaves as meiotic distorter/or selfish DNA. For exemple Wolbachia, a bacterial maternally transmitted endosymbiont in many arthropods, favors its own transmissiom through various tricks such as cytoplasmic incompatibilities, male feminizing , or even male killing.
What is the dynamics of these elements within the species, what are the impact on genome evolution?
Viruses: Besides these genetic entities, other intruders exist such as viruses. Viruses are not part of the genome. They arrive through infection of the cell, and are usually not transmitted to the progeny. Viruses are interesting candidates as vectors for horizontal transfer of TEs. Upon infection, TEs from the host can jump into the viral genome, and then benefit from both an easy means of escape and a route to travel to new species
Endogeneous viral elements (EVEs) are remnants of viral sequences are frequently found within genomes. They are transmitted to the next generation as genes and transposable elements. These EVEs are the signatures of past infections. Insertions DNA circles within host genomes has also been observed with a parasitoid domesticated virus, which helps suppress immunity of the larval host after parasitoid egg deposition.
Can TEs insert within viral genomes and be transmitted to new hosts? What determine the insertion of viral circles?
Interactions with the microbiome, interactions between host and parasites
Organisms do not live isolated in their environment but are permanently in interactions with other organisms. Close associations of eukaryotes with viruses and other microorganisms are frequent. These associated entities make up the microbiome. It can be intestinal but also on the skin, sometimes within the cells. This microbiome has a huge impact on the biology of the host. It helps for example in digestion, in producing vitamins or in protecting the host from pathogen attacks. The microbiome is highly dynamics.
How much does the microbiome composition change during development, with environmental changes or with diet? What is the involvment of the microbiome to the resilience of the species? Does the microbiome have a role in the success of invasive species?
Associations are sometimes parasitic. One organism develop detrimentally to the other. Cynipidae are wasps that lay eggs in plant tissues that will develop into a gall serving for protection and nutrition. The gall tissues or the developing larvae can themselves be parasitized by other Cynipidae wasps (inquilins or parasitoïds).
Evolution of gene networks
Each gene encodes a protein a with specific function but genes do not act isolated. They work within gene network. Furthermore, genes are often pleiotropic (they are involved in several functions) and can have epistatic effects. Genes can evolve by mutations affecting the coding sequences, but also by mutations affecting gene expression. Evolvability (the ability to evolve easily) is therefore constrained by the gene regulatory network. We draw models and run computer simulations to answer various questions about the evolution of gene networks. We anchor our models within biological examples.
How a mutation within a gene will impact the gene network? How robust and plastic are gene networks to mutations?
Adaptative and molecular evolution
Amylases are key enzymes involved in the degradation of complex sugars. Amylases genes are duplicated in the genomes of animals and perform tissue specific functions. Amyrel is a gene paralogous to the alpha-amylase, conserved in all muscomorphs. Its function is still unknown. It is expressed in the midgut of Drosophila. Compared to other amylases, it has the unique ability to produce glucose monomer.
Insects used as food
This research axis is a transverse axis in which all the teams in the lab are interested. Our model is the black soldier fly Hermetia illucens which is largely used as a source of proteins for feeding domesticated animals such as fishes. It is easy to rear and feed on organic waste. It has a worlwide distribution. Understanding its genetic diversity and phylogeography is a preliminary step before planning programs devoted to increase the productivity of the species.

