
Professeur Université Paris-Saclay
Co-Responsable équipe « Genomes et Evolution«
Téléphone +33 1 69 15 49 87
Courriel aurelie.hua-van@universite-paris-saclay.fr
Researcher ID: N-3495-2013
ORCID number: /0000-0001-7859-2788
Google Scholar Citation
Propositions de stage
Enseignements
- Transposable elements and evolution (Master)
- Biodiversité (L1)
- Animal and plant life cycle (L1)
- L’origine de la vie (Chimie-Biologie L1)
- Python (L2)
- Bioinformatique (L3)
Research Thematics
The core of my research topics is transposable elements (TEs, aka transposons), from an evolutionary perspective. They are basically selfish DNA present in every genome, with no obvious function for the cell. So, why are they here?, how did they get in?, why are they so numerous and diversified?, why can’t we get rid of them?, how do they evolve?, how do they interact with other genome components and with each other? What is the genome answer to these invaders?, what is the consequence of their presence in the genome?
We may have some answers to all these questions. But answers are always imprecise and always generate more questions…
For grabbing more informations on those genetic entities, I develop bioinformatics tools for analyzing TEs in sequenced genomes (mainly from eukaryots), I explore some theoretical frameworks using in silico simulations, and I run experimental evolution, using one of the best models for the biologist: the fruit fly Drosophila, and one of the best models for the « transposonist »: the mariner element (the historical one identified in 1985, a representative of Class II element)
By extension I am also interested with any other selfish DNA, and any interaction between genetic entities.
Dynamics and evolution of transposable elements
Transposable elements (TEs) usually amplify in genomes and populations from one initial copy, landed through horizontal transfer. It starts then to amplify, invading the genome, and the population, until selection or regulation counteract and stabilize the copy number. Transposable element copies accumulate mutations with time, and ultimately become inactive. Hence, a TE in a species is never forever active, but always leave tracks of its passage.
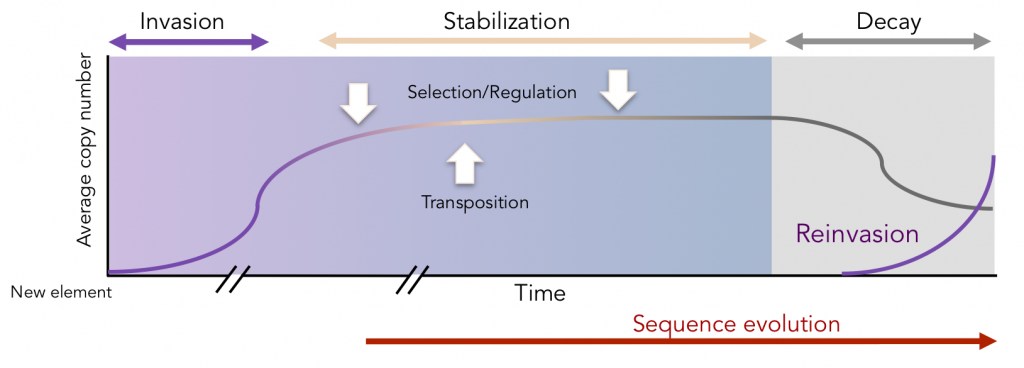
Within the genome, a lineage amplified from one copy is then monophyletic, and relationships between copies can be described by a phylogenetic analysis. if we suppose that copies accumulate mutations with time. Then the topology of the trees, and more particularly the deepness of the nodes can be used to estimate the past transposition rate, and its variation across time. The resulting graphs easily illustrate the evolutionary dynamic of a transposon family (Le Rouzic et al. 2013)
Eukaryotic Transposable Elements : Diversity, Structure and Classification
An update of the classification (Wicker et al 2007)
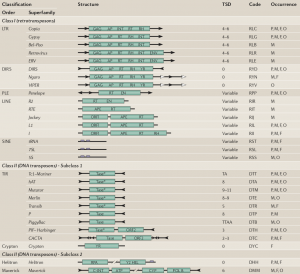
Transposable elements are typically divided into two main Classes. Class I corresponds to TEs that use an RNA intermediate for copy-and-paste transposition, whereas Class II transpose directly by excision and reinsertion (cut-and-paste mechanisms). Several subclasses and sub-families are then distinguished, according to their structural characteristics (presence of LTR, genes order…) and phylogenetic relationships (protein motifs)
The discovery of new atypic families of transposable elements has made necessary an update of the classification system. The classification proposed in 2007 by Wicker and coworkers is based on the classical one. A new criterion is used: the number of strand cut at the donor site during transposition defines two subclasses among the Class II. An intermediate hierarchical level is proposed : the Order, that relies on structural characteristics of elements
Transposases structure in the Mutator superfamily (Hua-Van and Capy, 2008)
Autonomous transposable elements encode one or several proteins that are required for transposition, for exemple an integrase, for Class I elements or a transposase for Class II elements. Both are involved in the reinsertion process and have endonucleases and strand transfer activities. In most families, those proteins belong to the Ribonuclease H-like large protein superfamily and are characterized by a catalytic motif DDE.
Mutator is a superfamily, comprising bacterial and eukaryotic elements. Bacterial Mutator families (IS256-like) contain a DDE atalytic domain, that differ for the classic DDE present in retroelement integrases. Notably, the two last residue are separated by more than 100 AA. In eukaryotic Mutator this motif is less conserved, and barely detectable. The comparison of the primary sequence of a large number of transposase, combined with secondary and tertiary structures analysis have allowed characterizing this catalytic motif. This illustrates the fact that for highly divergent protein, multi- level analyses are necessary .
Autonomous elements versus non-autonomous ones
The MITEs (Miniature Inverted-repeat Transposable Elements), and other non-autonomous elements, parasite the autonomous copies.
MITEs are non autonomous elements that uses the transposases encoded by related autonomous copies for multiply. This parasitic strategy is succesful and often lead to highly repeated MITE families. However newly formed MITEs that are not yet amplified are difficult to detect, because of their small size and the absence of coding regions. De novo detection of MITE in genomes relies mainly on their structural characteristic, i.e, the presence of terminal inverted repeats (TIRs) and the fact that they can transpose (at least two independent copies). Detection tools can only be efficient if they contain some methods to filter out the numerous false positive candidates.
Experimental evolution on Drosophila
Download AutomitAur.v1.0.1
Read Me ReadMe
The mariner transposable element
The Class II mariner family comprises numerous elements found in metazoa. All share a very homogeneous structure: About 1.3 kb long, they are characterized by two TIRs (Terminal Inverted Repeats) and one intronless ORF. Like all elements from the Tc1-mariner superfamily, they insert specifically into TA dinucleotide, and the transposase has a typical DDE catalytic motif (rve type).
mariner-like elements (MLEs) populate numerous metazoa genomes, and in particular Drosophila. The first mariner element was discovered in 1986 in Drosophila mauritiana, a sister species of Drosophila melanogaster. The active TE lineage is referred as mariner, but also bear alternative names (mos1, from the active copy), DROMAR1, Dmmar1 (in some database).
mariner in Drosophila genomes
The availability of hundreds sequenced Drosophila genomes offers the opportunity for deep comparative analysis.
The phenotypic excision assay
Somatic excision of mariner (type Mos1) can be visualised in Drosophila (D. melanogaster, D. mauritiana et D. simulans) using a phenotypic excision assay of the peach copy. This mariner copy is inserted in 5′ of the white gene, conferring flies a peach-colored-eyed phenotype [wpch or peach]. peach excision in eye cells restore white activity, leading to red spots or dots on a peach background. Such flies are said « mosaic ». This assay allows evaluating the influence various environmental or genomic factors on the (somatic) transposition level.

Photo: Fly with mosaic eyes, resulting from the excision of mariner from the white gene
This assay also offers a simple phenotypic way for following genome and population invasion by mariner. We follow experimental populations in which one mos1-carrying fly is introduced as a migrant, among 99 flies that only contain the inactive copy. At each generation, the extent of invasion can be followed by counting the proportion of mosaic flies. The copy number amplification is monitored by qPCR.
Publications
2021
Herpin A, Schartl M, Depincé A, Guiguen Y, Bobe J, Hua-Van A, Hayman ES, Octavera A, Yoshizaki G, Nichols KM, Goetz GW, Luckenbach JA. Genome Res. 2021 Allelic diversification after transposable element exaptation promoted gsdf as the master sex determining gene of sablefish. 31(8):1366-1380. doi: 10.1101/gr.274266.120.
Madi-Ravazzi L, Segala LF, Roman BE, Alevi KCC, Prediger C, Yassin A, Hua-Van A, Miller WJ. 2021 Integrative taxonomy and a new species description in the sturtevanti subgroup of the Drosophila saltans group (Diptera: Drosophilidae). Zootaxa. 4980(2):269292. doi: 10.11646/zootaxa.4980.2.3
Gauthier J, Boulain H, … , Huguet E, Herniou EA, Drezen JM. 2021. Chromosomal scale assembly of parasitic wasp genome reveals symbiotic virus colonization. Commun Biol. 4(1):104. doi: 10.1038/s42003-020-01623-8.
2020
Saint-Leandre B, Capy P, Hua-Van A, Filee J. 2020. piRNA and transposon dynamics in Drosophila: a female story. Genome Biology and Evolution. 12(6):931–947. doi :10.1093/gbe/evaa094
2019
Schneider DI, Ehrman L, Engl T, Kaltenpoth M, Hua-Van A, Le Rouzic A, Miller WJ. 2019. Symbiont-driven male mating success in the neotropical Drosophila paulistorum superspecies. Behavior genetics 49:83-98
Attardo GM, Abd-Alla AM, Acosta-Serrano A, Allen JE, Bateta R, Benoit JB, Bourtzis K, Caers J, Caljon G, Christensen MB. 2019. Comparative genomic analysis of six Glossina genomes, vectors of African trypanosomes. Genome biology 20:1-31
2018
Pereira CM, Stoffel TJR, Callegari-Jacques SM, Hua-Van A, Capy P, Loreto ELS. 2018. The somatic mobilization of transposable element mariner-Mos1 during the Drosophila lifespan and its biological consequences. Gene 679:65-72.doi: 10.1016/j.gene.2018.08.079
2017
Saint-Leandre B, Clavereau I, Hua-Van A, Capy P. 2017. Transcriptional polymorphism of piRNA regulatory genes underlies the mariner activity in Drosophila simulans testes. Mol Ecol 26:3715-3731.doi: 10.1111/mec.14145
Bouallegue M, Rouault JD, Hua-Van A, Makni M, Capy P. 2017. Molecular evolution of piggyBac superfamily: from selfishness to domestication. Genome Biol Evol evw292 [pii]
2016
2015 and before
- Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, Fiston-Lavier AS, Hua-Van A, Hubley R, Kapusta A, et al. 2015. A call for benchmarking transposable element annotation methods. Mob DNA 6:13.doi: 10.1186/s13100-015-0044-6
- Gasmi L, Boulain H, Gauthier J, Hua-Van A, Musset K, Jakubowska AK, Aury JM, Volkoff AN, Huguet E, Herrero S, et al. 2015. Recurrent Domestication by Lepidoptera of Genes from Their Parasites Mediated by Bracoviruses. PLoS Genet 11:e1005470.doi: 10.1371/journal.pgen.1005470
- Wallau GL, Capy P, Loreto E, Hua-Van A. 2014. Genomic landscape and evolutionary dynamics of mariner transposable elements within the Drosophila genus. BMC Genomics 15:727.doi: 10.1186/1471-2164-15-727
- Le Rouzic A, Payen T, Hua-Van A. 2013. Reconstructing the evolutionary history of transposable elements. Genome Biol Evol 5:77-86.doi: 10.1093/gbe/evs130
- Kamoun C, Payen T, Hua-Van A, Filee J. 2013. Improving prokaryotic transposable elements identification using a combination of de novo and profile HMM methods. BMC Genomics 14:700.doi: 10.1186/1471-2164-14-700
- Da Lage JL, Binder M, Hua-Van A, Janecek S, Casane D. 2013. Gene make-up: rapid and massive intron gains after horizontal transfer of a bacterial alpha-amylase gene to Basidiomycetes. BMC Evol Biol 13:40.doi: 10.1186/1471-2148-13-40
- Fort P, Albertini A, Hua-Van A, Berthomieu A, Roche S, Delsuc F, Pasteur N, Capy P, Gaudin Y, Weill M. 2012. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol Biol Evol 29:381-390.doi: 10.1093/molbev/msr226
- Wallau GL, Hua-Van A, Capy P, Loreto EL. 2011. The evolutionary history of mariner-like elements in Neotropical drosophilids. Genetica 139:327-338.doi: 10.1007/s10709-011-9552-6
- Hua-Van A, Le Rouzic A, Boutin TS, Filee J, Capy P. 2011. The struggle for life of the genome’s selfish architects. Biol Direct 6:19.doi: 10.1186/1745-6150-6-19
- Hua-Van A, Le Rouzic A, Boutin TS, Filee J, Capy P. 2011. The struggle for life of the genome’s selfish architects. Biol Direct 6:19.doi: 10.1186/1745-6150-6-19
- Dufresne M, Lespinet O, Daboussi MJ, Hua-Van A. 2011. Genome-wide comparative analysis of pogo-like transposable elements in different Fusarium species. J Mol Evol 73:230-243.doi: 10.1007/s00239-011-9472-1
- Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367-373.doi: 10.1038/nature08850
- Rouault JD, Casse N, Chenais B, Hua-Van A, Filee J, Capy P. 2009. Automatic classification within families of transposable elements: application to the mariner Family. Gene 448:227-232.doi: 10.1016/j.gene.2009.08.009
- Picot S, Wallau GL, Loreto EL, Heredia FO, Hua-Van A, Capy P. 2008. The mariner transposable element in natural populations of Drosophila simulans. Heredity (Edinb) 101:53-59.doi: 10.1038/hdy.2008.27
- Hua-Van A, Capy P. 2008. Analysis of the DDE motif in the Mutator superfamily. J Mol Evol 67:670-681.doi: 10.1007/s00239-008-9178-1
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973-982.doi: nrg2165 [pii]
- Maisonhaute C, Ogereau D, Hua-Van A, Capy P. 2007. Amplification of the 1731 LTR retrotransposon in Drosophila melanogaster cultured cells: origin of neocopies and impact on the genome. Gene 393:116-126.doi: S0378-1119(07)00075-3 [pii]
- Dufresne M, Hua-Van A, El Wahab HA, Ben M’Barek S, Vasnier C, Teysset L, Kema GH, Daboussi MJ. 2007. Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics 175:441-452.doi: genetics.106.064360 []
10.1534/genetics.106.064360 - Hua-Van A, Le Rouzic A, Maisonhaute C, Capy P. 2005. Abundance, distribution and dynamics of retrotransposable elements and transposons: similarities and differences. Cytogenet Genome Res 110:426-440.doi: 84975
- Lobo NF, Hua-Van A, Li X, Nolen BM, Fraser MJ, Jr. 2002. Germ line transformation of the yellow fever mosquito, Aedes aegypti, mediated by transpositional insertion of a piggyBac vector. Insect Mol Biol 11:133-139.doi: 317 [pii]
- Hua-Van A, Langin T, Daboussi MJ. 2002. Aberrant transposition of a Tc1-mariner element, impala, in the fungus Fusarium oxysporum. Mol Genet Genomics 267:79-87.doi: 10.1007/s00438-002-0638-9
- Villalba F, Lebrun MH, Hua-Van A, Daboussi MJ, Grosjean-Cournoyer MC. 2001. Transposon impala, a novel tool for gene tagging in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 14:308-315.doi: 10.1094/MPMI.2001.14.3.308
- Lobo N, Li X, Hua-Van A, Fraser MJ, Jr. 2001. Mobility of the piggyBac transposon in embryos of the vectors of Dengue fever (Aedes albopictus) and La Crosse encephalitis (Ae. triseriatus). Mol Genet Genomics 265:66-71
- Li Destri Nicosia MG, Brocard-Masson C, Demais S, Hua Van A, Daboussi MJ, Scazzocchio C. 2001. Heterologous transposition in Aspergillus nidulans. Mol Microbiol 39:1330-1344.doi: mmi2323 [pii]
- Hua-Van A, Pamphile JA, Langin T, Daboussi MJ. 2001. Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol Gen Genet 264:724-731
- Hua-Van A, Langin T, Daboussi MJ. 2001. Evolutionary history of the impala transposon in Fusarium oxysporum. Mol Biol Evol 18:1959-1969
- Hua-Van A, Daviere JM, Kaper F, Langin T, Daboussi MJ. 2000. Genome organization in Fusarium oxysporum: clusters of class II transposons. Curr Genet 37:339-347
- Hua-Van A, Hericourt F, Capy P, Daboussi MJ, Langin T. 1998. Three highly divergent subfamilies of the impala transposable element coexist in the genome of the fungus Fusarium oxysporum. Mol Gen Genet 259:354-362
